The coronavirus (COVID-19) was identified in Wuhan province of China towards the end of 2019. Since then, it has spread rapidly across the world. We can simulate the outbreak in NetLogo, to analyse the gravity of the spread, well in advance.
According to different studies, the coronavirus can survive on a surface for up to 72 hours. This has quite literally locked down the world. The virus, which can spread through person to person contact, has infected over a million (as of the first week of April) and led to the deaths of thousands around the world. Even though modern medicine has advanced a lot, we have no antidote for such new viral diseases. We can’t even predict how long developing a vaccine might take, and whether that may be too late for many.
To be well prepared for such viral outbreaks, it is important to know how the disease will spread, well in advance. Each country has the data on its population, density, demographics, etc. It is possible to simulate an outbreak for different communities/countries if we quantify the parameters properly in a simulation. Netlogo is software that can simulate such scenarios on a large scale. It is GNU GPL licensed free software, designed by Uri Wilensky, who was the director of Northwestern University in 1999. It is a JVM based cross-platform multi-agent simulation platform.
NetLogo doesn’t require coding knowledge or any other prerequisites. It is designed for a broad audience. So, anybody from secondary school students to graduates can make use of this software for their field of interest. An inbuilt model library has various models available on disease spread. Some are explained below.
Disease Solo
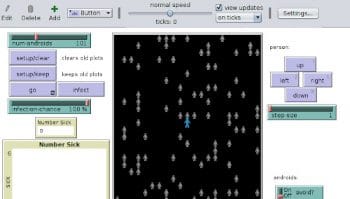
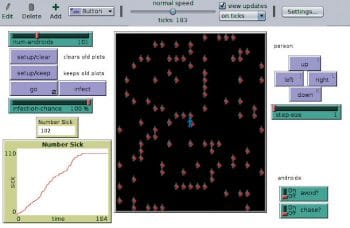
Disease Solo can simulate the spread of an outbreak within a population. One agent in population is controlled manually, while others are controlled by the computer. Figure 1 shows the initial situation of a single infected person among a 100 people. After 183 steps, all 101 people are infected, as shown in Figure 2. The graph in Figure 2 looks similar to how the outbreak happened in China.
epiDEM
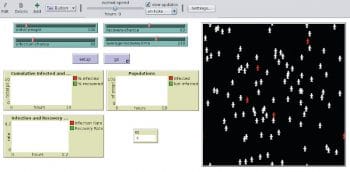
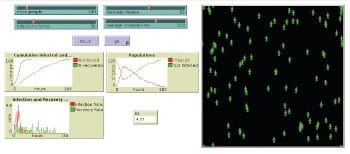
The Epidemiology Understanding Disease Dynamics and Emergence through Modeling (epiDEM) basically simulates the spread of an infectious disease in a closed population (community/country). Systemic dynamics infection from one person to a susceptible population can be studied. It has an additional feature of gauging the recovery rate. Figure 3 shows how, initially, some people were infected in a population of 100. After 184 steps, Figure 4 shows a recovery rate of 60 per cent.
epiDEM Travel and Control
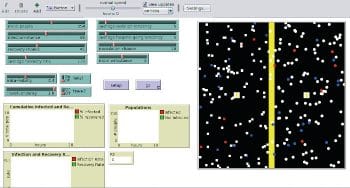
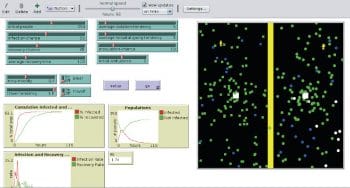
epiDEM Travel and Control is an extension of the basic model. This models the spread of the disease in a semi-closed population, but with additional features that factor in travel, isolation, quarantine, inoculation, and links between individuals. However, it is assumed that the virus does not mutate and that complete recovery is possible. After 93 hours, Figure 6 shows that people have recovered, compared to Figure 5. It is possible to study the outbreak of a disease within a population in a community or area, with all the available parameters.
There are other models available with various features that include simulating the spread of the disease in a number of different conditions and environments, the interactions of agents, etc. However, we stop here. Netlogo can be installed and simulated with Ubuntu-14.04 LTS OS.