The entry of open source tools in the life sciences arena has proven to be a boon. Open source tools can be used in the predictive and diagnostic fields to provide better medical treatment. Through their use in brain mapping and DNA studies, open source tools can even be used to combat crime.
Nowadays, the applications of information and communications technology (ICT) are not limited to data transmission, cloud deployments, social media, Web servers and mobile applications. Since the last decade, IT is touching every area of the social and corporate world including health and medical sciences. Most of the medical diagnosis laboratories are now equipped with advanced computerised machines to accurately diagnose and fetch the parameters of the human body. These diagnostic machines include those used for magnetic resonance imaging (MRI), computed tomography (CT), electroencephalography (EEG), etc. These systems provide a higher degree of accuracy in the analysis of the human body, assisting doctors in diagnosing the disease and thus recommending a suitable course of treatment.
In addition to diagnostic machines, software tools and libraries are also used. These software tools and applications evaluate the biological data collected from the computerised diagnostic machines. Thus, the concept of bioinformatics has evolved, which uses software tools and applications to understand the biological and medical data. These software suites make use of high performance programming languages at the back-end to process and evaluate the biological data set, leading to effective treatment.
Bioinformatics is the interdisciplinary area that integrates biology, computer science, mathematics, engineering, chemistry and statistics for advanced predictions and analytics. The field of molecular biology is also closely associated with bioinformatics for accurate analysis of biological structures. Molecular biology deals with the deep analysis of the bimolecular movements in the cells of the body along with the details of proteins, DNA, RNA and biosynthesis.
Data sets for research in medicine and biology
With the deployment of computerised machines, researchers in diagnostic and medical sciences are taking assistance from software professionals in their field so that the programming modules can be processed by these developers. Even computer scientists are now taking the interdisciplinary field of bioinformatics for their research so that their programming knowledge can be utilised for the health sciences.
There are numerous medical data sets available for research, which are released by the diagnostic laboratories so that the overall architecture and structure of medico-biological data can be analysed by software experts. The programmers working in bioinformatics can download these medical data sets and they can perform the analysis using effective algorithms.
The software tools that can be used for the analysis and evaluation of medical data for specific types of data sets are summarised below.
OpenEEG (http://openeeg.sourceforge.net/doc/)
OpenEEG is free and open source software that can be used for EEG signal analysis with numerous libraries as add-ons, including Neuroserver, BioEra, BrainBay, Brainathlon, BrainWave Viewer and EEGMIR.
EEGNET (https://sites.google.com/site/eegnetworks/)
This is a free and open source tool for the analysis and visualisation of EEG brain signals. It has features to visualise the brain network.
BioSig (http://biosig.sourceforge.net/)
BioSig is a software library under free and open source distribution with many features of biomedical signal processing. This library has excellent features to process biosignals including electrocorticogram (ECoG), electromyogram (EMG), electrocardiogram (ECG), electrooculogram (EOG), electroencephalogram (EEG), respiration and many others. In addition, the interfacing toolboxes and drivers for Octave, MATLAB, Python, PHP, Perl, Ruby, Tcl, C and C++ are also available. The key areas of brain-computer interfaces, psychology, neuroinformatics, cardiovascular systems, neurophysiology and sleep research are effectively processed in BioSig.
GenomeTools (http://genometools.org/)
GenomeTools is open source software for the analysis of genome and biological parameters. It has a free library of tools for bioinformatics. The APIs in C are available with detailed manuals. In addition, the deep analysis of biological structures is integrated in GenomeTools.
Working with BioPython for molecular biology (http://biopython.org/)
BioPython provides the set of tools and libraries for the analysis and computation of biological structures. It is available for free and open source distribution and is promoted by the Open Bioinformatics Foundation (OBF). It can parse the files of bioinformatics into the data structures that can be processed by Python code.
The following international formats are supported in BioPython:
- UniGene
- PubMed
- GenBank
- Medline
- GenBank
- FASTA
- Clustalw
- Blast
Use the following command to install BioPython in Ubuntu:
<span style="color: #000000;">$ sudo apt-get install python-biopython</span> |
The following command will install the documentation along with BioPython:
<span style="color: #000000;">$ sudo apt-get install python-biopython-doc</span> |
BioSQL (http://biosql.org) can be used with BioPython to store a biological database. To integrate BioSQL, the following instruction is executed:
<span style="color: #000000;">$ sudo apt-get install python-biopython-sql</span> |
Sequence is the key object in bioinformatics. Sequences can be processed in BioPython with the following instructions:
<span style="color: #000000;">>>> from Bio.Seq import Seq>>> my_seq = Seq(“MyDefinedSequence”)>>> my_seqSeq(‘ MyDefinedSequence ‘, Alphabet())>>> print(my_seq)MyDefinedSequence>>> my_seq.alphabetAlphabet()</span> |
Complement and reverse complement
These are very simple — the methods return a new Seq object with the appropriate sequence and the same alphabet, as shown below:
<span style="color: #000000;">>>> from Bio.Seq import Seq>>> from Bio.Alphabet import generic_dna>>> my_values_dna = Seq(“MY_VALUES_DNA”, generic_dna)>>> my_values_dnaSeq(‘MY_VALUES_DNA’, DNAAlphabet())>>> my_values_dna.complement()Seq(‘ATCATGTGACCA’, DNAAlphabet())>>> my_values_dna.reverse_complement()Seq(‘ACCAGTGTACTA’, DNAAlphabet())</span> |
Transcription functions on DNA and RNA
If you have a DNA sequence, you may want to turn it into RNA. In bioinformatics we normally assume the DNA is the coding strand (not the template strand); so this is a simple matter of replacing all the thymines with uracil:
<span style="color: #000000;">>>> my_values_dnaSeq(‘MY_VALUES_DNA’, DNAAlphabet())>>> my_values_dna.transcribe()Seq(‘AGUACACUGGU’, RNAAlphabet())</span> |
With the specification of RNA, the associated DNA can be fetched:
<span style="color: #000000;">>>> my_values_rna = my_values_dna.transcribe()>>> my_values_rnaSeq(‘AGUACACUGGU’, RNAAlphabet())>>> my_values_rna.back_transcribe()Seq(‘MY_VALUES_DNA’, DNAAlphabet())>>> my_values_rnaSeq(‘AGUACACUGGU’, RNAAlphabet())>>> my_values_rna.back_transcribe().reverse_complement()Seq(‘ACCAGTGTACT’, DNAAlphabet())</span> |
Sleep EEG analysis in GNU Octave
Assorted signals are delivered to all parts of the body so that the other organs can communicate with each other for specific or general purposes. One of the key signals in the human brain is electroencephalography (EEG), which is generated from the brain, even when asleep or unconscious. Electroencephalography (EEG) signals comprise brain waves that can be evaluated using GNU Octave. The analysis on sleeping disorders and various diseases can be done with EEG evaluation.
GNU Octave (https://www.gnu.org/software/octave/) is one of the powerful and multi-functional tools used for engineering and scientific applications of research. The simulations related to engineering as well as medicine can be implemented with the assorted tool boxes and functions in Octave. It is used as an effective alternate to MATLAB since it is open source and can be freely distributed. A number of tool boxes for different applications are available in GNU Octave, which can be used for optimisation and predictive analysis.
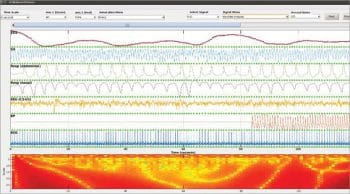
The Wave Form Database (WFDB) package can be integrated with GNU Octave. This package is equipped with the functions and modules for EEG and brain signal evaluations. A similar process is followed in case of brain mapping or brain fingerprinting for criminal investigation when the subject is in an unconscious state. There are assorted stages of sleep or unconscious states which can be analysed from EEG signals after recording from the electrodes. This process assists in the forensic analysis of the person while in the unconscious state. By this evaluation, the medical disorders can also be detected using the WFDB package in Octave. The following are the excerpts of Benchmark Sleep Stages which can be evaluated using the WFDB package in GNU Octave so that the overall state of the nervous system can be evaluated and predictions made, along with diagnosing brain disorders.
Stage 1: Tiredness, drowsiness, the pre-sleep stage and lethargy
- Eye activities
- Rolling eye movements
- Sharp transients
Stage 2: Normal night sleep
- Sleep spindles
- Slow eye movement
Stage 3: Delta sleep or slow wave sleep
- Sleep time of 6.5 hours
With the following instruction, the demonstration of the WFDB tool box can be viewed in Octave.
<span style="color: #000000;">>> wfdbdemo</span> |
The following instructions can be executed to read and plot the ECG signal from the data set repository of PhysioBank:
<span style="color: #000000;">[time,signal]=rdsamp(‘mitdb/100’,1);plot(time,signal);</span> |
(Source: https://www.physionet.org/physiotools/matlab/wfdb-app-matlab/)
Using similar methodology, the waveform of arterial blood pressure (ABP) can be analysed using the wabp function.
Scope of research in biomedical engineering
Nowadays, bioinformatics and biomedical predictive analytics are two key domains of research for assorted applications. The extraction, processing and predictive mining from the brain, heart and other human body generated signals are evaluated with the use of information technology. The data sets from Physionet, UCSD, FPMS and others can be used for the research work in bioinformatics with the integration of data mining and machine learning tools.